We study the mechanisms of intracellular protein degradation to solve problems of biological and clinical importance.
Theme 1. Elucidating the Mechanism of Allosteric Regulation of Mycobacterial Proteases
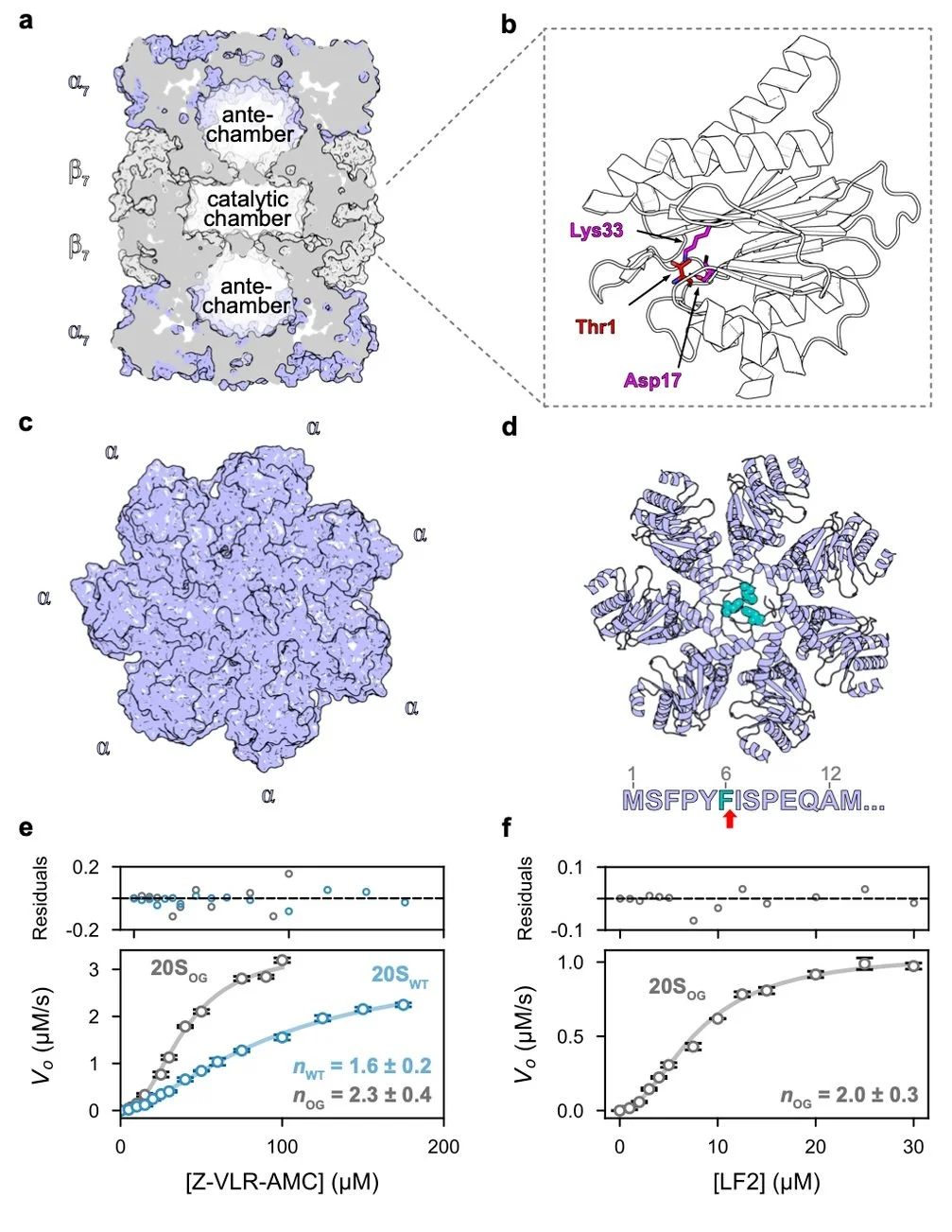
A key focus of our group is the structural and functional dissection of the Mycobacterium tuberculosis (Mtb) proteasome system, a critical machinery for the pathogen's intracellular survival and immune evasion. With rising multidrug-resistant (MDR) TB cases, there is an urgent need for mechanistically guided therapeutic strategies. We investigate the allosteric regulation of the Mtb 20S proteasome and its associated activators using cryo-EM, NMR, and HDX-MS, integrated with biochemical and cell-based assays to uncover long-range communication within the proteasome machinery.
Our work has led to multiple structural breakthroughs. We were the first to establish the structural basis of Mtb 20S core particle long-range allosteric regulation [Turner et al., Nat. Commun. 2025], structural insights into substrate engagement by a bacterial proteasome activator [Davis et al., under review], and the dynamic behaviou of substrate-tagging enzymes [Plourde et al., JBC 2025]. This research is funded by a CIHR Project Grant.
Our work has been featured in News & Views highlights and recognized through New Investigator awards and keynote invitations. This theme continues to drive major advances and collaborative opportunities, setting the stage for future discoveries and therapeutic development.
Theme 2. Mechanism of Allosteric Inhibition and Activation of Human Mitochondrial Proteases
Our second major research area explores the mitochondrial ClpP protease, a serine protease essential for mitochondrial proteostasis and a therapeutic target in acute myeloid leukemia (AML). AML cells rely heavily on mitochondrial metabolism, and ClpP inhibition offers a selective vulnerability. We partner with Dr. Aaron Schimmer (PMCC), Dr. Natalie Zeytuni (McGill), and Dr. Rima Al-Awar (OICR) to identify allosteric inhibitors of ClpP using NMR, HDX-MS, cryo-EM, and biochemical assays.
Our contributions include the discovery that serine phosphorylation (pSer) acts as a degradation signal in mitochondrial ClpXP [Feng et al., PNAS 2025], structural elucidation of ClpP activation by active-site inhibitors [Goncalves et al., PNAS 2025], and the development of first-in-class nanomolar allosteric inhibitors targeting a newly defined hotspot [Goncalves et al., under review]. These compounds demonstrated efficacy in AML models, paving the way for clinical translation. This research is funded by a CIHR Project Grant, and a Cancer Research Society grant.
We are now expanding into mitochondrial LONP1, another AAA+ protease implicated in AML. Preliminary data on LONP1 regulation and inhibitor binding supports a CIHR resubmission focused on this target. This program firmly positions our group at the forefront of mitochondrial protease biology and cancer therapeutics.
Theme 3. Advancing Mass Spectrometry Tools to Probe Biomolecular Structure and Dynamics
Our third research theme focuses on developing and applying mass spectrometry-based tools to interrogate protein conformational dynamics. Building on my PhD work with Lars Konermann, we have applied HDX-MS, radical footprinting, and electrospray ionization studies to challenging systems such as FoF1-ATP synthase [Vahidi et al., PNAS 2016], membrane proteins, and megadalton complexes.
In our lab, we introduced neutral-pH fluorinated ethylamines for native MS [Davis et al., Anal. Chem. 2023] and improved HDX-MS workflows for large complexes [Vosper et al., JASMS 2025]. While method development is a smaller part of our program, it directly enhances our capabilities and benefits the broader community.
We collaborate with Prof. Evan Williams (UC Berkeley) on charge detection mass spectrometry (CDMS) and partner with Waters Corporation developing the applications of ELIT-based CDMS technology. This research is supported by an NSERC Discovery Grant.